YesWorkflow Annotation Example
YesWorkflow is a tool that models conventional scripts and exposes the underlying workflow view (prospective provenance). Firstly, a YesWorkflow user can add special YesWorkflow (YW) comments to existing scripts. These comments declare how data are used and results are produced, step by step, by the script. Then, the YesWorkflow tool interprets the YW comments and produces a graphical output that reveals the computation steps and the data flows hidden in the script. The YesWorkflow keyword definitions are listed at the table below. Lastly, YesWorkflow provides query capability for the prospective and retrospective provenance of the scripts. We use Alice’s soil mapping script to demonstrate the usage of the YesWorkflow tool.
| Keywords | Definitions |
| @BEGIN <name_of_code_block> | Specifies the beginning of a code block. Each @BEGIN annotation must be paired with an @END annotation later in the same source file. A @BEGIN-@END pair can occur between other pairs to indicate nested code blocks |
| @END <name_of_code_block> | Specifies the end of a code block. @END must be paired with a @BEGIN annotation earlier in the same source file. A @BEGIN-@END pair can occur between other pairs to indicate nested code blocks |
| @IN <input_data_name>[@URI <path_to_file | uri_template>] | Specifies relevant input data elements of a code block. @IN should appear in the code bracketed by associated @BEGIN and @END tags |
| @PARAM <parameter_name>[@URI <path_to_file | uri_template>] | Specifies a block’s parameters. It defines the values that control how the input data are processed. @PARAM should appear in the code bracketed by associated @BEGIN and @END tags |
| @OUT <output_data_name>[@URI <path_to_file | uri_template>] | Models relevant output data elements of a code block. @OUT should appear in the code bracketed by associated @BEGIN and @END tags |
| @AS <alias_name> | Links a program variable in the script code to the scientist’s concepts |
| @URI <path_to_file | uri_template> | Declares the path to a resource. A URI template can be either a path to a file or a template having template variables for multiple files (or other data resource) written or read by a script in a specific code block. |
| @RET <returned_var>[@URI <path_to_file | uri_template>] | Defines returned results at the conceptual level |
| @CALL <function_name> | Specifies a function call |
| @DESC <description_string> | Specifies a description to a program or a port |
| @FILE <path_to_file> | Specifies a path to a local file |
| @LOG <log_file_name> | Specifies a log file |
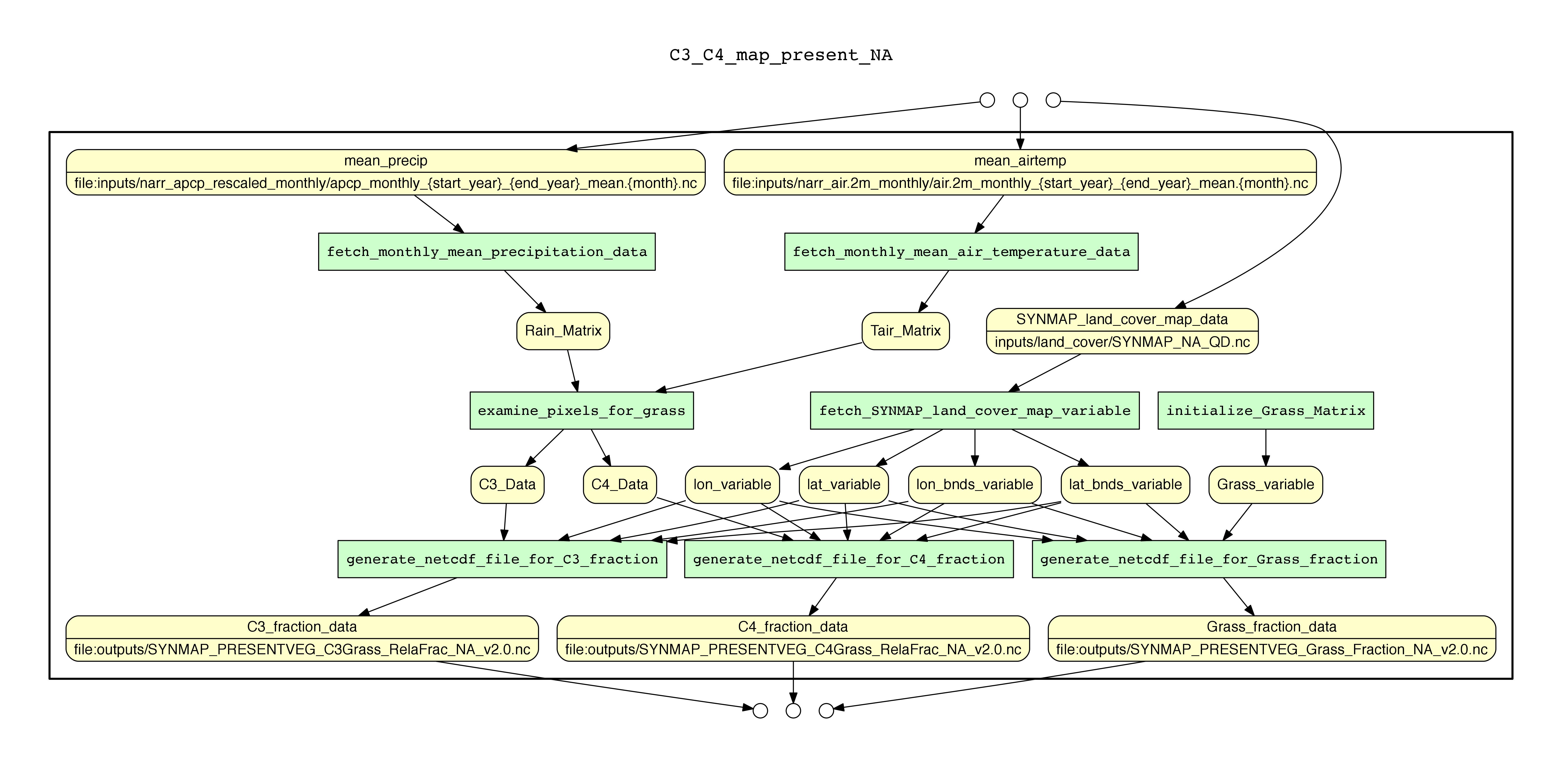
%% @begin fetch_SYNMAP_land_cover_map_variable
% @in mstmip_SYNMAP_NA_QD.nc @as SYNMAP_land_cover_map_data
% @out lon @as lon_variable
% @out lat @as lat_variable
% @out lon_bnds @as lon_bnds_variable
% @out lat_bnds @as lat_bnds_variable
%% Load input: SYNMAP land cover classification map;
%% also read coordinate variables to re-use them later
grass_type=[19,20,21,22,23,24,25,26,27,38,41,42,43];
sncid=netcdf.open('inputs/land_cover/SYNMAP_NA_QD.nc', 'NC_NOWRITE');
fvid=netcdf.inqVarID(sncid, 'biome_frac');
frac=netcdf.getVar(sncid,fvid);
tvid=netcdf.inqVarID(sncid, 'biome_type');
type=netcdf.getVar(sncid,tvid);
lon_vid=netcdf.inqVarID(sncid, 'lon');
lon=netcdf.getVar(sncid,lon_vid);
lat_vid=netcdf.inqVarID(sncid, 'lat');
lat=netcdf.getVar(sncid,lat_vid);
lon_bnds_vid=netcdf.inqVarID(sncid, 'lon_bnds');
lon_bnds=netcdf.getVar(sncid,lon_bnds_vid);
lat_bnds_vid=netcdf.inqVarID(sncid, 'lat_bnds');
lat_bnds=netcdf.getVar(sncid,lat_bnds_vid);
netcdf.close(sncid)
%% @end fetch_SYNMAP_land_cover_map_variable